16S rRNA Microbiology Sequence Analysis protocol
1. Go to 16S_Microbiology_Sequencing_Data and download the .ab1 file that matches your sequence tube code.
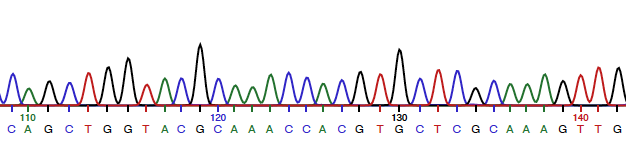
2. Open the .ab1 file with ApE. See whether you have a "good" read. You should see that most of it has something like this.
1. Go to 16S_Microbiology_Sequencing_Data and download the .ab1 file that matches your sequence tube code.
2. Open the .ab1 file with ApE. See whether you have a "good" read. You should see that most of it has something like this.
3. If you have a "bad" read with your seq file full of N's (not just at the ends, but all over), and an equally bad chromatogram, go to 16S good reads and pick a new prefix and files.
4. Use File -> "New DNA from basecalls". This opens a window with the sequence
5. Highlight and copy the DNA sequence.
6. Go to BLASTn
7. Paste your sequence into the Query Sequence window.
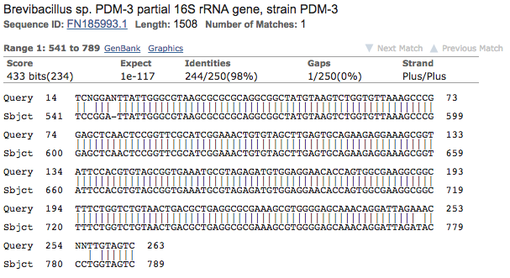
8. Click the BLAST button. After the website is finished aligning, a new page will appear. Go to "Alignments" to see the sequence alignment:
9. What bacterial species gives the best match?
10. Click on the link following "Sequence ID," Is there anything noteworthy about the species or sample your sequence matched with (there might not be anything interesting ... use your judgement!)
11. Record the data in the 16S Data spreadsheet with the instructor.
a. Your name
b. Your sample (from the microbiology plate)
c. Your sequence code (or "bad")
d. The species (example: Brevibacillus sp. PDM-3)
e. The Expect value (example: 1e-117)
f. The percent identity (example: 98%)
g. Any noteworthy facts
4. Use File -> "New DNA from basecalls". This opens a window with the sequence
5. Highlight and copy the DNA sequence.
6. Go to BLASTn
7. Paste your sequence into the Query Sequence window.
8. Click the BLAST button. After the website is finished aligning, a new page will appear. Go to "Alignments" to see the sequence alignment:
9. What bacterial species gives the best match?
10. Click on the link following "Sequence ID," Is there anything noteworthy about the species or sample your sequence matched with (there might not be anything interesting ... use your judgement!)
11. Record the data in the 16S Data spreadsheet with the instructor.
a. Your name
b. Your sample (from the microbiology plate)
c. Your sequence code (or "bad")
d. The species (example: Brevibacillus sp. PDM-3)
e. The Expect value (example: 1e-117)
f. The percent identity (example: 98%)
g. Any noteworthy facts